Talk to Our Scientists
"*" indicates required fields
Why CovalX Epitope Mapping services will help your project?
Epitope mapping has become one of the key elements of both vaccine and drug development. Epitope mapping is the process of identifying the binding site of a protein to its corresponding antigen. FDA and EMEA guidelines recommend molecular analysis of the interaction site between a drug and its target for regulatory filing. In addition, a better understanding of the interaction site is useful to develop more potent drugs and to protect products with patents.
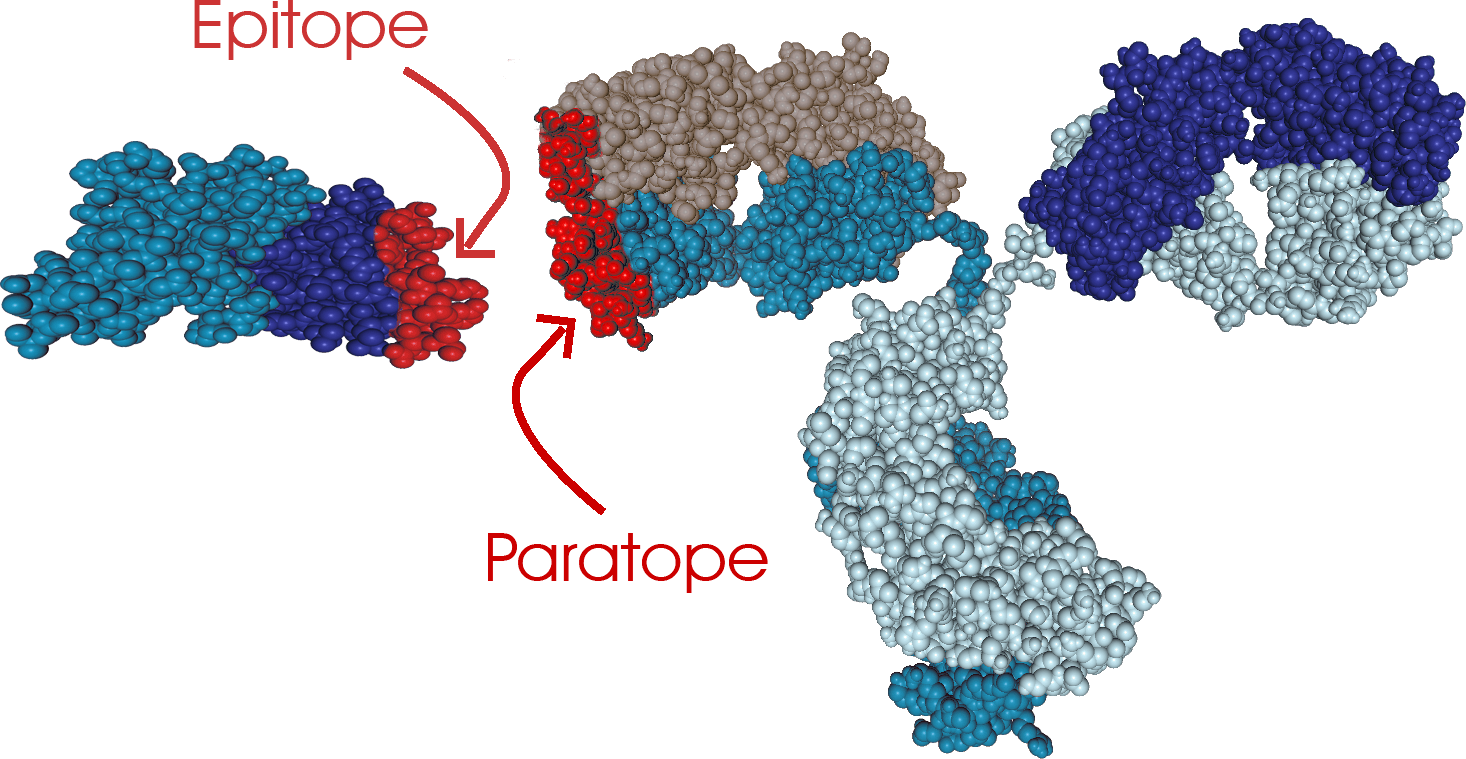
Epitope mapping is the process of identifying the binding site(s) on an antigen to its corresponding antibody and is a key element for vaccine and drug development. Over a decade, CovalX’s epitope mapping services have been successfully utilized for more than 700 mAbs with a success rate of over 95% and providing well-supported data for use in patents for companies of all sizes.
 CovalX Epitope Mapping
CovalX Epitope Mapping Why Epitope Mapping by Mass Spectrometry?
Mass spectrometry is one of the key elements for drug development. At every stage of development, from the early stage to quality control, mass spec technology allows the characterization of the drug with the highest confidence and reproducibility.
CovalX is utilizing this key mass spec technology for epitope mapping. Based on our unique high-mass technologies for measuring intact protein complexes combined with high-resolution instrumentation, CovalX is providing unparalleled services.
Which Epitope Mapping Techniques Offer CovalX Technology?
CovalX offers two techniques for conformational epitope mapping by mass spectrometry: Hydrogen Deuterium eXchange (HDX-MS) and Crosslinking Mass Spectrometry (XL-MS). Each of these technologies provides unique advantages.
CovalX Epitope Mapping Techniques
Hydrogen Deuterium eXchange
(HDX-MS)
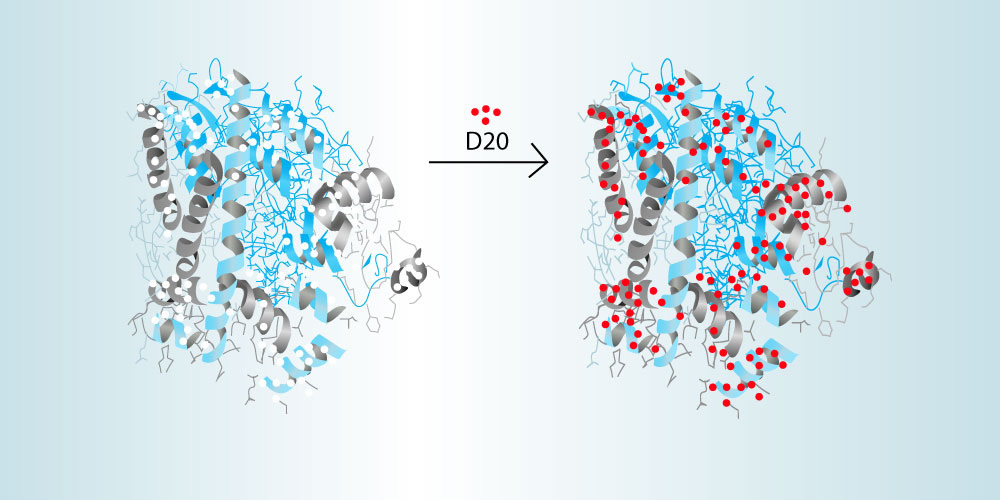
HDX-MS measures the accessibility of hydrogen molecules in the backbone of a protein structure. During analysis, both the unbound antigen and the bound antibody-antigen complex are incubated in deuterated (D2O) water in order to exchange any amide hydrogens with deuterium from exposed amino acids of the protein’s backbone. The location of these deuterium molecules on the proteins sequence can then be carefully determined using high-resolution mass spectrometry. By comparing the unbound antigen with the bound antibody-antigen complex, the residues of the epitope can be determined. This technique requires careful control of the temperature, pH, and time of the reactions. Because of this, CovalX uses highly optimized using modern robotics and software packages designed specifically for HDX analysis. CovalX is offering this technique through our contract research services facility. More information can be seen here.
Other applications of HDX-MS take advantage of a high-resolution molecular structural view through CovalX’s HDX analysis for the following projects:
- Biosimilar characterization
- Higher order structure
- Conformational/Protein dynamics
- Small molecules interaction
- Protein-Protein interactions
- Protein folding characterization
Crosslinking coupled Mass Spectrometry
(XL-MS)
XL-MS begins by binding the antibody and the antigen with a mass-labeled chemical crosslinker. Next, the presence of the complex is confirmed using high-mass MALDI detection. Since after applying the crosslinking chemistry, the Ab/Ag complex is extremely stable, multiple enzymes (five utilized in parallel) and digestion conditions can be applied to the complex to provide many different overlapping peptides. Identification of these peptides is performed using high-resolution Orbitrap™ mass spectrometry and MS/MS techniques. The identification of the crosslinked peptides is determined using mass tags linked to the crosslinking reagents. After MS/MS fragmentation and data analysis using specific interaction software(s), both epitope and paratope are determined in the same experiment. Because of the high sensitivity and accuracy of mass spectrometry detection, very little quantity of material is required. CovalX is offering this technique through our contract research services facility. More information can be seen here.
Other applications of XL-MS. CovalX unique XL protocol will let you succeed on projects like:
- Protein-Protein Interactions
- Mapping Aggregates
- Parallel Paratope Mapping Analysis
 Coval-X linear conformational epitope mapping service
Coval-X linear conformational epitope mapping service Linear Vs Conformational Epitopes
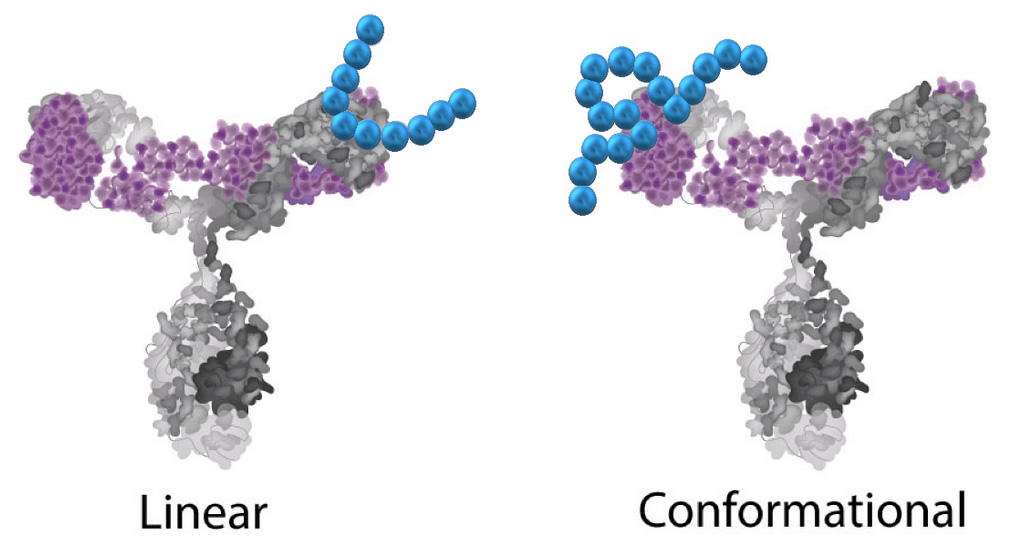
Epitopes can be defined by two general categories, linear or conformational. A linear (or sequential) epitope is composed of a linear stretch of amino acids in the sequence which has no 3-dimensional structure. A conformational epitope is one that requires tertiary folding to create the proper binding region.
Conformational epitopes are most often found in regions that have broken (discontinuous) in the amino acid binding sequence requiring the structure to be folded to bring the regions into proximity. Conformational mapping techniques can be used for identifying either linear or conformational epitopes. However, linear techniques cannot map conformational epitopes.
Depending on the “resolution” of the technique utilized, the region identified can be individual amino acids, small groups of amino acids, or peptide stretches of the protein. In general, higher resolution techniques are more complex, time-consuming, and require more material/time.
How CovalX Helps Ensure Success
One of the unique capabilities at CovalX is the rapid and robust ability to detect large molecules and intact protein complexes by MALDI mass spec. For all epitope mapping projects, individual proteins are first screened to confirm mass and detect significant multimerization. Then the complex is detected intact, and stoichiometry is determined before epitope mapping is performed.
This rapid screening has proven to be beneficial in saving our customers time and money prior to conducting the more advanced and costly epitope mapping projects. The cost and time of this process are standard in all project quotes for both XL-MS and HDX-MS epitope mapping projects.