Talk to Our Scientists
"*" indicates required fields
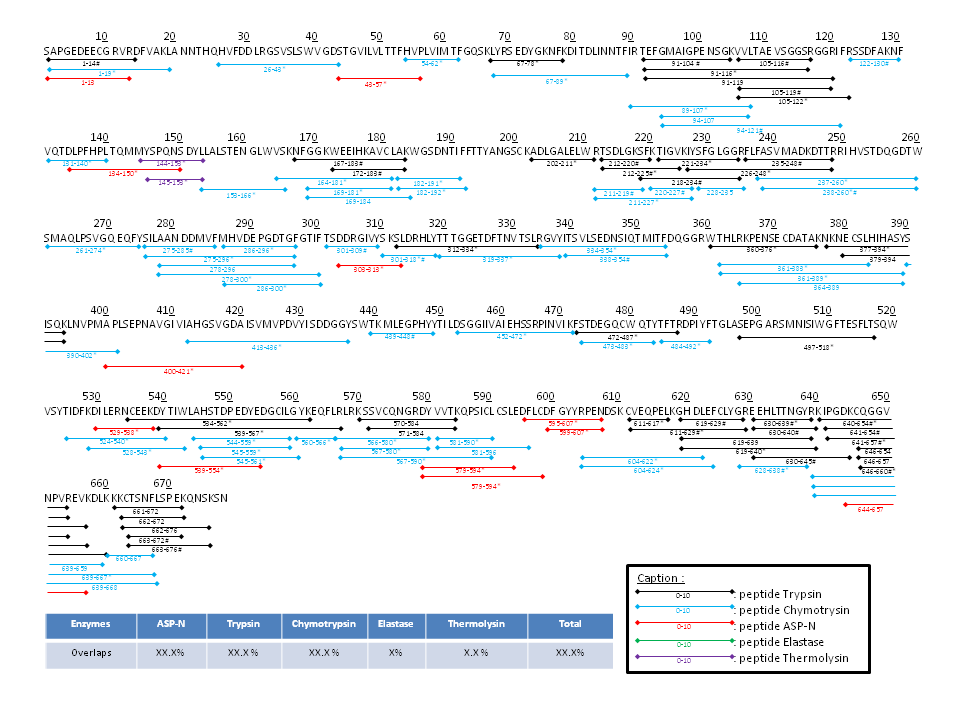
Sequence verification by overlapping peptide identification
Proteins are digested with multiple enzymes run in parallel (5 enzymes shown below) and the resulting peptides are analyzed by high-resolution nano-Liquid Chromatography-Mass Spec (nLC-MS/MS). The peptides are fragmented inside the mass spectrometer to produce patterns which are then matched to the protein sequence using specialized computer software.
Simple sample buffers such as water or volatile compounds are compatible with direct in-solution digestion. However, more complex buffers such as salts, detergent, salts or glycerol may require an additional clean-up step prior to sample analysis. The sensitivity of this technique can be 1 ng or less starting material for a given protein.
A full report is generated containing instrument specifications and variables and showing coverage of the different peptides color coded across the sequence of the protein, as shown in image.
- Sample types: 2D gel spots, SDS-PAGE bands, recombinant proteins, purified proteins
- Data turn around: Two weeks or less