Talk to Our Scientists
"*" indicates required fields
Sequence optimization for manufacturing
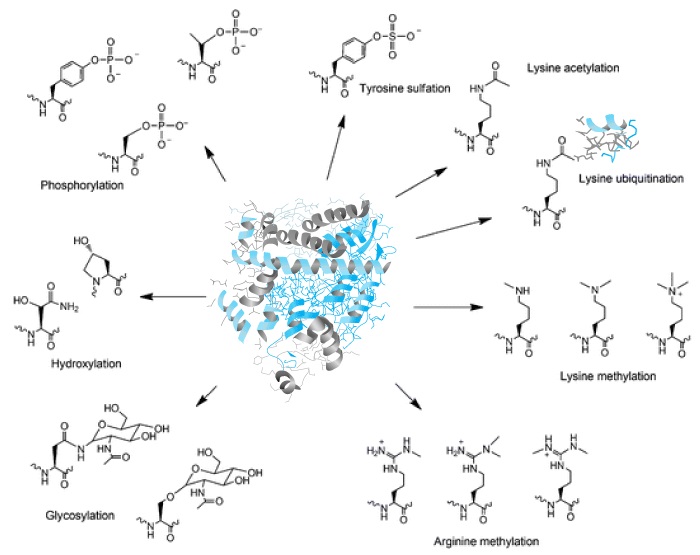
This service is used to detect protein post-translational modifications (PTM). The service can be applied to purified proteins, recombinant proteins or proteins in complex mixtures. Samples can be submitted as purified proteins or isolated such as from gel bands. The range of modifications are limited to those readily identifiable in a database search, such as methylation of lysine or arginine, ubiquitination of lysine, deamidation of glutamine or asparagine, glycation of lysine, phosphorylation of serine, threonine or tyrosine (without enrichment), or acetylation of lysine.
In more complex samples, fractionation can be employed. The extent of the fractionation correlates with the number of proteins and their PTMs that are detected. The greater the number of fractions, the greater the number of PTMs that can be identified.
Following digestions with multiple enzymes, the peptide mixtures are analyzed by high-resolution LC/MS/MS. The peptides are fragmented in the mass spectrometer to yield diagnostic patterns that can be matched to protein sequence databases via Mascot software analysis.
- Sample types: 2D gel spots, SDS-PAGE bands, recombinant proteins, purified proteins
- Data turn around: Three weeks or less