Talk to Our Scientists
"*" indicates required fields
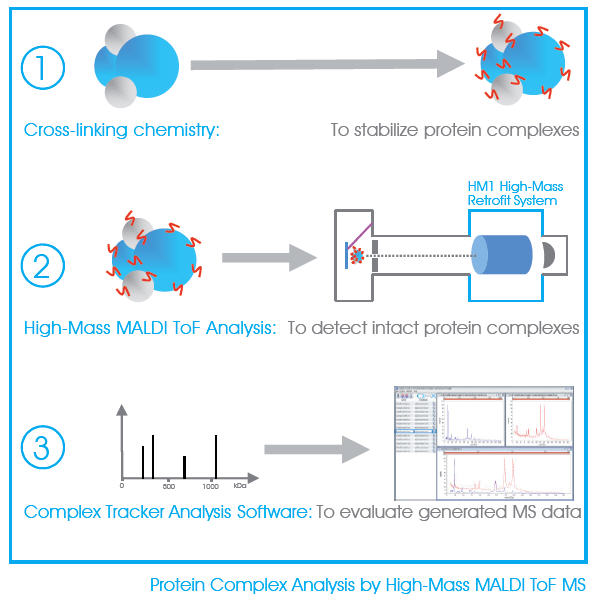
CovalX has developed and optimized a method for a fast, sensitive and accurate analysis of protein complexes directly by High-Mass MALDI mass spectrometry. Unfragmented and undigested, the protein complexes are detected intact using specially developed cross-linking reagents and high-mass detection system. There is no sample pre-labeling or tagging required and no immobilization, the samples are measured directly in solution.
Using cross-linking chemistry and High-Mass MALDI ToF mass spectrometry, it becomes possible to easily analyze non-covalent protein complexes in the 1-2000 kDa range. The sample to be analyzed can be a simple aggregate multimers (dimer, trimer, etc), antibody fragment (Fab: one-to-one), monoclonal (two-to-one), or more advanced stoichiometry of nearly any protein-protein interaction.
The use of High-Mass MALDI allows a much faster, cheaper and more accurate method for studying protein interactions:
- The stoichiometry of the complex: unique information for understanding the shape of your protein complex.
- Clear identification of the partners involved in the complex.
- No need for immobilization, the complex is detected without labeling.
- Fast: 30 minutes per assay.
- Specific: only the specific complexes are detected.
- Sensitive: Only a few hundred femtomoles of the protein sample is a few µl are required to achieve the analysis.
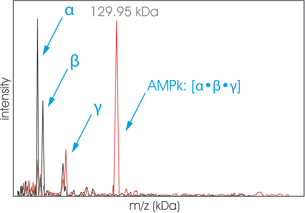
In the spectrum presented, the protein complex AMPk is analyzed by High-Mass MALDI ToF mass spectrometry before cross-linking (black) and after cross-linking (red) with CovalX’s K200 MALDI MS Analysis kit. (Contact Us to learn more about this Kit) Before cross-linking, the three subunits α, β, and γ of the protein complex are detected with m=30.22 kDa, m=37.38 kDa, and m=64.97 kDa. After cross-linking, the intact protein complex corresponding to the stoichiometry [α·β·γ] is detected with m=129.95 kDa. Riek U et al.; J. Biol. Chem.; 2008.
CovalX services then use our Complex Tracker software for the internal correction and direct overlay and subtraction analysis of the control and complex spectra providing a rapid and accurate characterization of the complex.
 CovalX Complex Analysis
CovalX Complex Analysis CovalX Biotherapeutic Lab Services
CovalX Biotherapeutic Lab Services