Talk to Our Scientists
"*" indicates required fields
Why Choose CovalX’s Epitope Mapping Services by XL-MS (Crosslinking Mass Spectrometry)?
CovalX offers a unique analytical service for epitope mapping based on our patented technology. Our service allows conformation or linear epitope determination of monoclonal antibodies targeting any type of protein antigen (No limit on molecular weight).
 CovalX Epitope Mapping
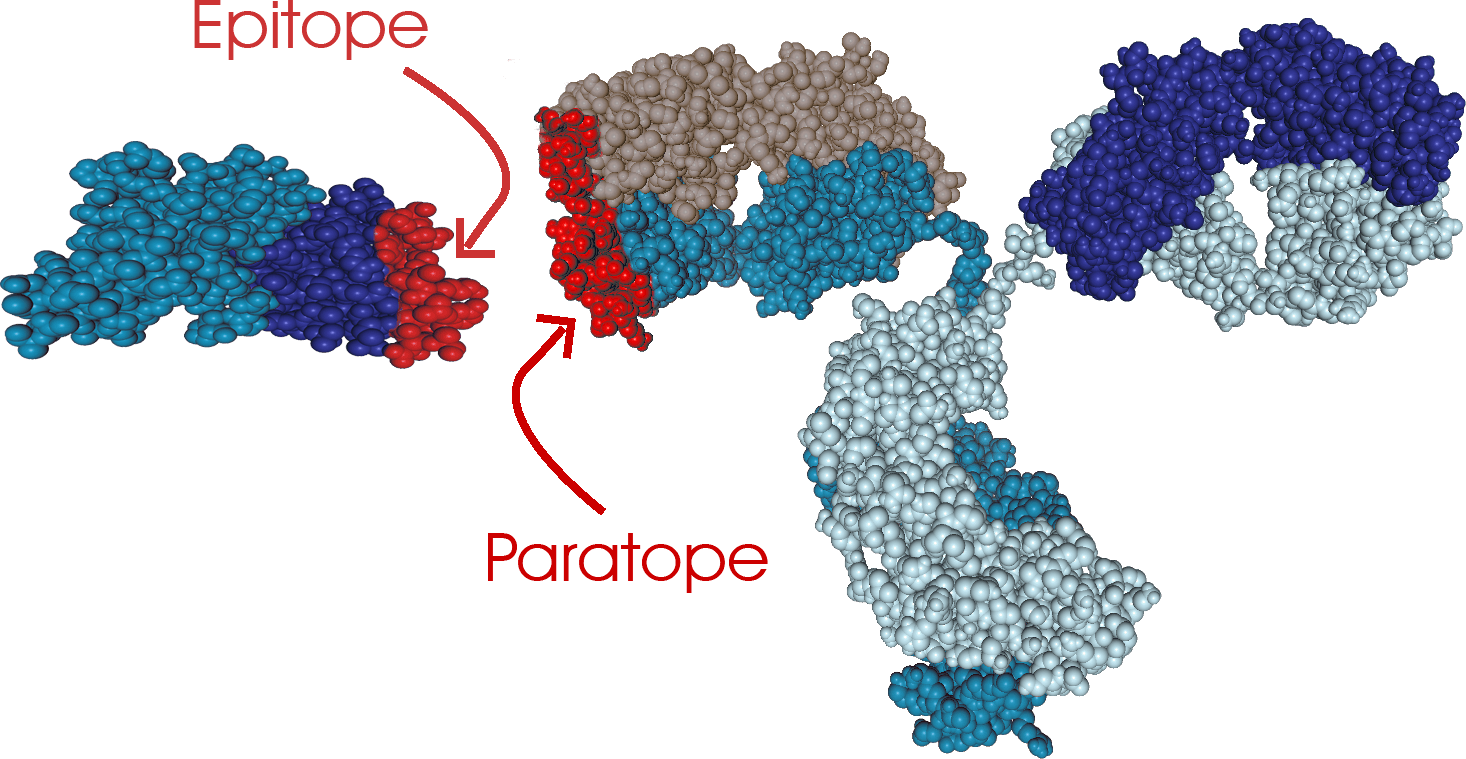
CovalX Epitope Mapping Based on mass spectrometry, our epitope mapping services allow epitope characterization with high resolution in fast scheduled turnaround time. The use of mass spectrometry combined with cross-linking gives unique information on the structure of the antibody-antigen interactions, including paratope identification in every analysis. In addition, our methods allow direct insight such as stoichiometry of the interaction (mono- or bi-valency), aggregation and integrity of the antibody.
Why is CovalX’s Epitope Mapping Services the best Option?
CovalX offers two techniques for conformational epitope mapping by mass spectrometry: Hydrogen Deuterium eXchange (HDX-MS) and Crosslinking Mass Spectrometry (XL-MS). Each of these technologies provides unique advantages.
| Readout | Characterization of conformational or linear epitopes |
|---|---|
| Determination of both epitope and paratope | |
| Affordable | Competitive cost compared to other mapping techniques |
| Low Sample Consumption | Only 200μg of each antigen/antibody required |
| Full Compatibility | No need for immobilization, recombinant protein work, peptide synthesis or micro-array. The interaction is analyzed directly by Mass Spectrometry in solution. Analysis of full mAbs, fAbs, tagged proteins or modified proteins. |
| Fast Turnaround | Four to five weeks delivery time (less with RUSH) |
| Quality Work | Expert team with more than 15+ years of epitope mapping experience using mass spectrometry. |
CovalX Epitope Mapping by XL-MS Step by Step
Based on our unique High Mass MALDI mass spectrometry technology and decades of crosslinking experiences, CovalX offers a sensitive characterization of an epitope with high resolution.
Using mass spectrometry and covalent tagging (though crosslinking), it is possible to characterize conformational and linear epitopes in weeks with a resolution of 3-4 amino acids or less. Without the need for peptide synthesis or micro-array chips, mass spectrometry allows for the identification of the interacting peptides with unmatched accuracy.
This technique provides the lowest cost, lowest sample consumption and shortest analysis time possible when compared with other conformational epitope mapping techniques.
Additionally, because the protein complexes are first covalently stabilized, multiple enzymes can be utilized providing highly optimized digestion conditions with the highest sequence coverage possible.
Step 1: Initial Sample Screening
Before the high-resolution analysis of the epitope and paratope can begin, CovalX performs High-Mass MALDI analysis on the antibody, antigen and the intact antibody/antigen complex.
The goals of these analyses are to verify:
- The integrity of both the antibody and the antigen
- The possible aggregation of the antibody
- The possible multimerization of the antigen
- The stoichiometry of the antibody/antigen interaction (monovalency, bivalency, etc)
- Verification that the specific intact cross-link antibody/antigen complex is present in the sample before proteolytic cleavage
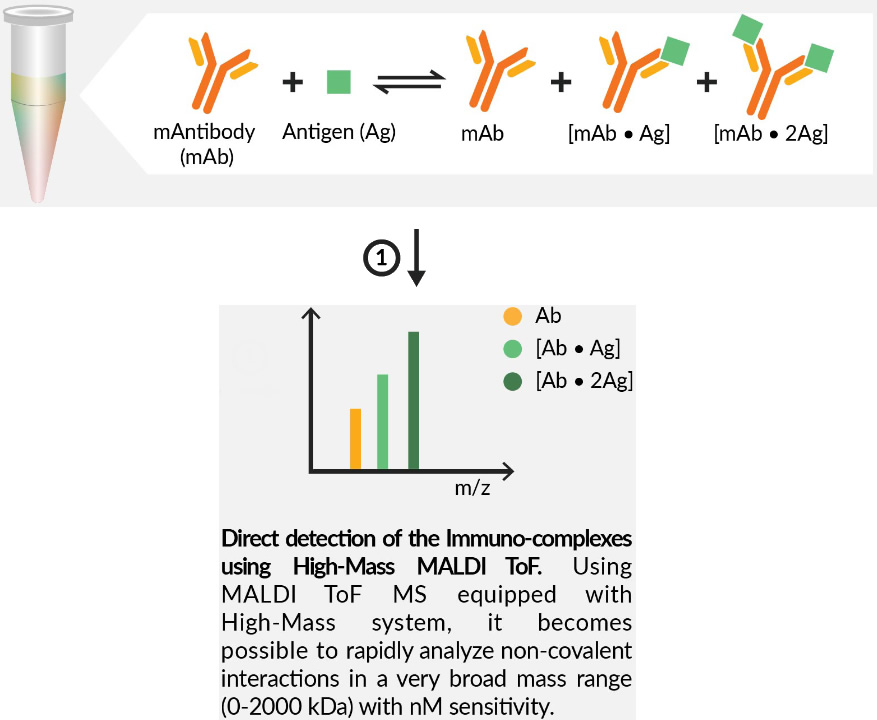
Step 2: Crosslinking Mass Spec Mapping (XL-MS)
After initial sample screening is completed, an individual length of cross-linker is selected which is shown to effectively stabilize the complex using our high mass MALDI detection. A 50:50 mixture of deuterated to undeuterated cross-linker is created to provide a unique mass tag for detecting the linkers location within the proteins sequence. CovalX uses unique software to later detect these deuterated cross-linkers directly, allowing the highest degree of confidence in the data.
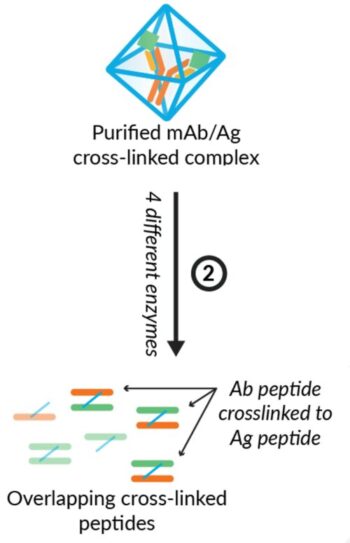
Step 3: Multiple Enzymes Provide Highest Coverage
After stabilization, four different enzymes are utilized in parallel for four separate and comprehensive digestions. Trypsin, Chymotrypsin, Elastase, and Thermolyse are each utilized individually to ensure the most thorough detection possible. The peptide coverage from the sum of these digestions provides the highest sequence coverage available with the most overlapping possibilities.
Finally, the peptide data is matched to the sequences using dedicated software to ensure proper identification of both the paratope and epitope from one data set.

Talk to Our Scientists
Our team is available to answer any of your questions about our products and services
Timeline, Sample Requirements & Resolution
- Timeline: 4-5 weeks for one epitope (less with RUSH); 5-6 weeks for projects with up to 5 epitopes against the same antigen.
- Sample Requirement: 200 μg of antigen and 200 μg of mAb/epitope.
- Resolution down to 3-4 amino acids or less.
- Applicable for conformational, discontinuous and linear epitopes.
- Applicable for any therapeutic proteins or antigen. No limit on size. No limit in complexity (multimers, hydrophobicity)
Download supporting materials
 CovalX Crosslinking Mass Spec Mapping (XL-MS) Brochure
CovalX Crosslinking Mass Spec Mapping (XL-MS) Brochure
For example patents, publications, or comparison data please see our library or contact us directly.
An overview of epitope mapping technologies can be seen here.